Selected publications (full list)
We are committed to open access. If you find a manuscript that we have co-authored but is not available here, please contact us directly and we will be happy to send you an electronic copy.
* Stands for first co-authorship
# Stands for last co-authorship
EGFR/FOXO3a/BIM signaling pathway determines chemosensitivity of BMP4-differentiated glioma stem cells to temozolomide
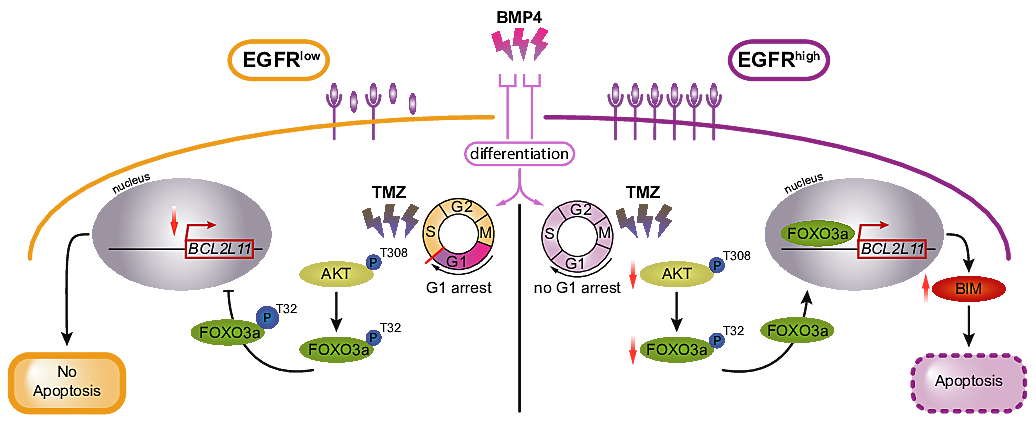
Accumulating evidence suggests that glioma stem cells (GSCs), which are rare cells characterized by pluripotency and self-renewal ability, are responsible for glioblastoma (GBM) propagation, recurrence and resistance to therapies. Bone morphogenic proteins (BMPs) induce GSC differentiation, which leads to elimination of GSCs and sensitization of glioma to chemotherapeutics. Alterations in the epidermal growth factor receptor (EGFR) gene are detected in more than half of GBMs; however, the role of EGFR in the chemoresistance of GSCs remains unknown. Here, we examined whether EGFR signaling affects BMP4-induced differentiation of GSCs and their response to the alkylating drug temozolomide (TMZ). We show that BMP4 triggers the SMAD signaling cascade in GSCs independent of the EGFR level. BMP4 downregulated the levels of pluripotency markers (SOX2 and OLIG2) with a concomitant induction of an astrocytic marker (GFAP) and a neuronal marker (β-Tubulin III). However, GSCs with different EGFR levels responded differently to treatments. BMP4-induced differentiation did not enhance sensitivity to TMZ in EGFRlow GSCs, in contrast to EGFRhigh GSCs, which underwent apoptosis. We then identified differences in cell cycle regulation. In EGFRlow cells, BMP4-triggered G1 cell cycle arrest which was not detected in EGFRhigh cells. RNA-seq profiles further highlighted transcriptomic alterations and distinct processes characterizing EGFR-dependent responses in the course of BMP4-induced differentiation. We found that the control of BIM (the pro-apoptotic BCL-2 family protein) by the AKT/FOXO3a axis only operated in BMP4-differentiated EGFRhigh cells upon TMZ treatment.
Paper: Exp Mol Med. 2020 Aug;52(8):1326-1340. doi: 10.1038/s12276-020-0479-9 | Pubmed | PDF
Mapping chromatin accessibility and active regulatory elements reveals new pathological mechanisms in human gliomas
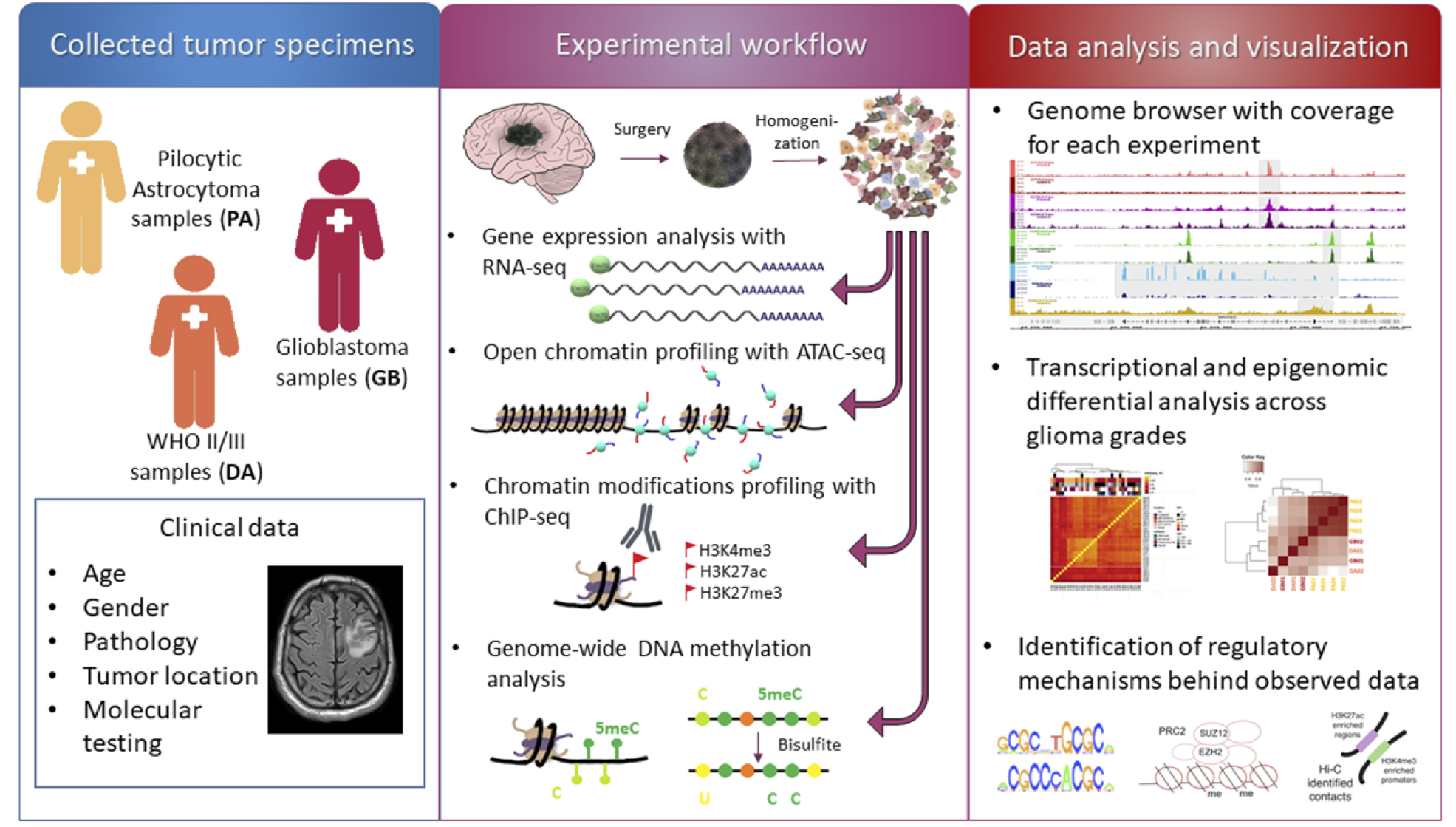
Chromatin structure and accessibility, and combinatorial binding of transcription factors to regulatory elements in genomic DNA control transcription. Genetic variations in genes encoding histones, epigenetics-related enzymes or modifiers affect chromatin structure/dynamics and result in alterations in gene expression contributing to cancer development or progression. Gliomas are brain tumors frequently associated with epigenetics-related gene deregulation. We performed whole-genome mapping of chromatin accessibility, histone modifications, DNA methylation patterns and transcriptome analysis simultaneously in multiple tumor samples to unravel novel epigenetic dysfunctions driving gliomagenesis. Based on the results of the integrative analysis of the acquired profiles, we created an atlas of active enhancers and promoters in benign and malignant gliomas. We explored these elements and intersected with Hi-C data to uncover molecular mechanisms instructing gene expression in gliomas.
Paper: bioRxiv | PDF
Down-regulation of IKKβ expression in glioma-infiltrating microglia/macrophages is associated with defective inflammatory/immune gene responses in glioblastoma
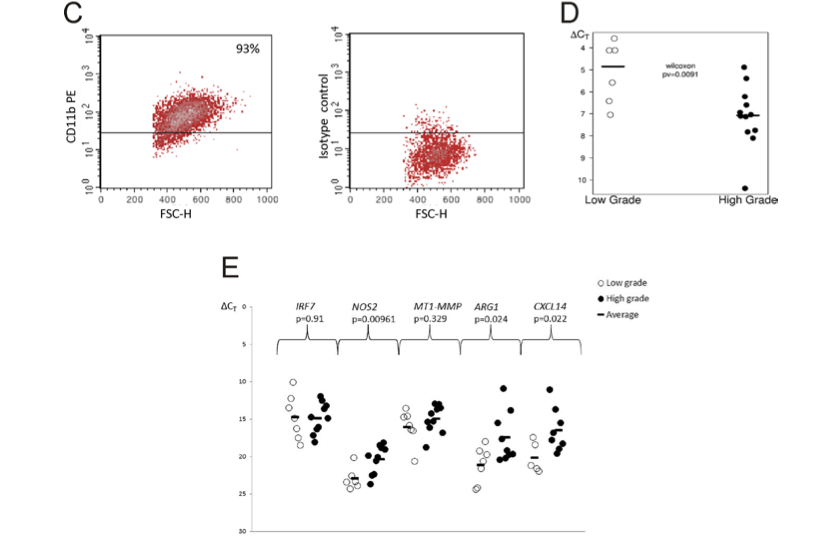
Glioblastoma (GBM) is an aggressive malignancy associated with profound host immunosuppression. Microglia and macrophages infiltrating GBM acquire the pro-tumorigenic, M2 phenotype and support tumor invasion, proliferation, survival, angiogenesis and block immune responses both locally and systematically. Mechanisms responsible for immunological deficits in GBM patients are poorly understood. We analyzed immune/inflammatory gene expression in five datasets of low and high grade gliomas, and performed Gene Ontology and signaling pathway analyses to identify defective transcriptional responses. The expression of many immune/inflammatory response and TLR signaling pathway genes was reduced in high grade gliomas compared to low grade gliomas. In particular, we found the reduced expression of the IKBKB, a gene coding for IKKβ, which phosphorylates IκB proteins and represents a convergence point for most signal transduction pathways leading to NFκB activation. The reduced IKBKB expression and IKKβ levels in GBM tissues were demonstrated by qPCR, Western blotting and immunohistochemistry. The IKKβ expression was down-regulated in microglia/macrophages infiltrating glioblastoma. NFκB activation, prominent in microglia/macrophages infiltrating low grade gliomas, was reduced in microglia/macrophages in glioblastoma tissues. Down-regulation of IKBKB expression and NFκB signaling in microglia/macrophages infiltrating glioblastoma correlates with defective expression of immune/inflammatory genes and M2 polarization that may result in the global impairment of anti-tumor immune responses in glioblastoma.
Paper: Oncotarget. 2015 Oct 20;6(32):33077-90. doi:10.18632/oncotarget.5310 | Pubmed | PDF